1: Department of Biomedical Engineering, Duke University, Durham NC, USA.
2: Department of Electrical and Computer Engineering, Duke University, Durham NC, USA.
Standard microscopes offer a variety of settings to help improve the visibility of different specimens to the end microscope user. Increasingly, however, digital microscopes are used to capture images for automated interpretation by computer algorithms (e.g., for feature classification, detection or segmentation), often without any human involvement. In this work, we investigate an approach to jointly optimize multiple microscope settings, together with a classification network, for improved performance with such automated tasks. We explore the interplay between optimization of programmable illumination and pupil transmission, using experimentally imaged blood smears for automated malaria parasite detection, to show that multi-element “learned sensing” outperforms its single-element counterpart. While not necessarily ideal for human interpretation, the network’s resulting low-resolution microscope images (20X-comparable) offer a machine learning network sufficient contrast to match the classification performance of corresponding high-resolution imagery (100X-comparable), pointing a path towards accurate automation over large fields-of-view.
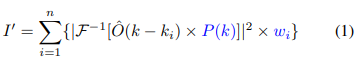
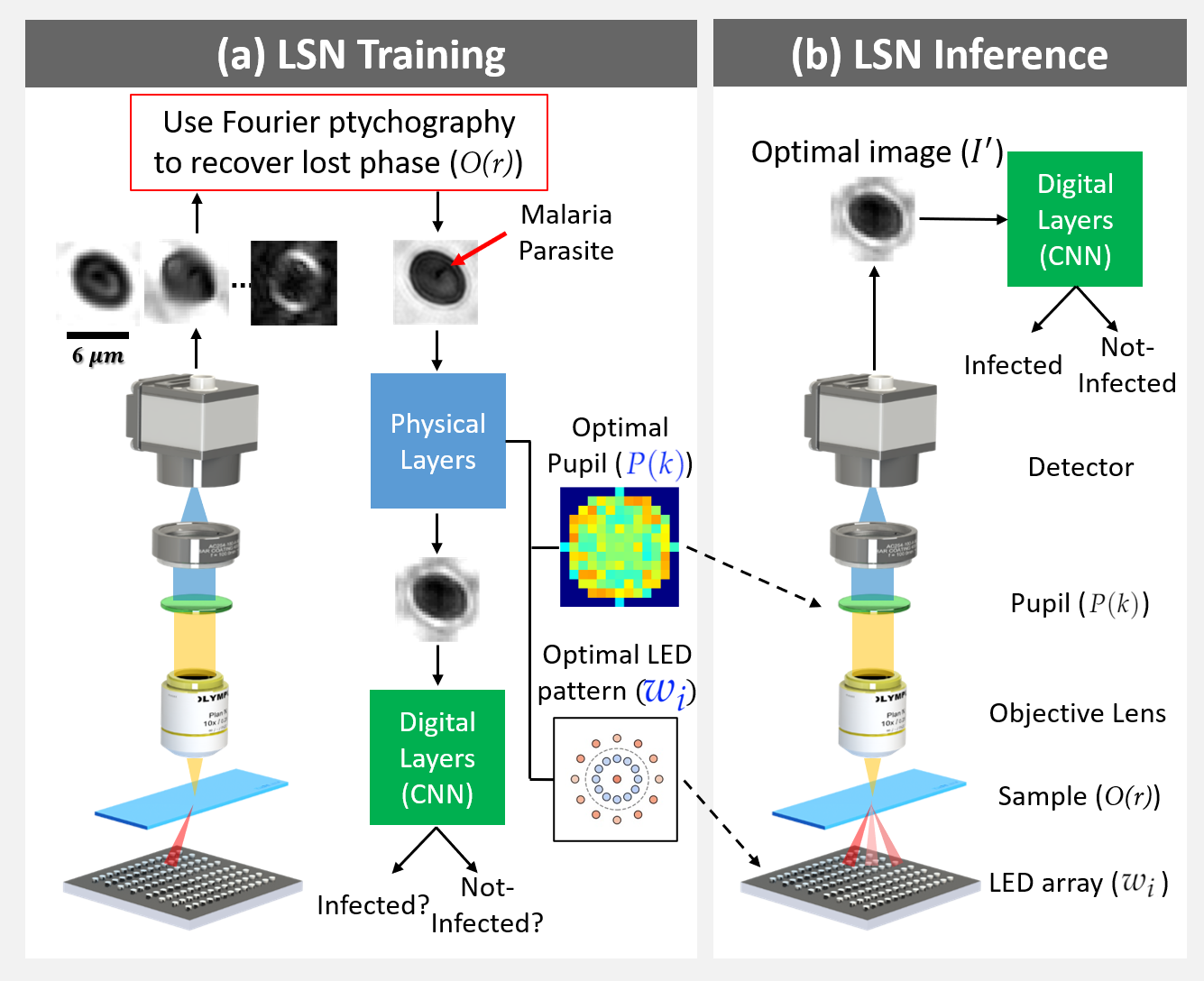
Fig. 1: Learned sensing network (LSN) framework: (a) During training, LEDs are turned on time-sequentially to capture multiple images, which are then processed with Fourier ptychography to recover the lost phase. Reconstructions are sent through the LSN physical layers (Eq. 1), facilitating the optimization of pupil transmission and illumination weights, culminating in an optimized image for LSN digital layer processing. Supervised learning is used to optimize both physical and digital layer weights. (b) During inference, the optimized pupil transmission and LED illumination pattern are used to generate the optimal image at the detector, which directly enters the digital CNN layers for accurate classification.
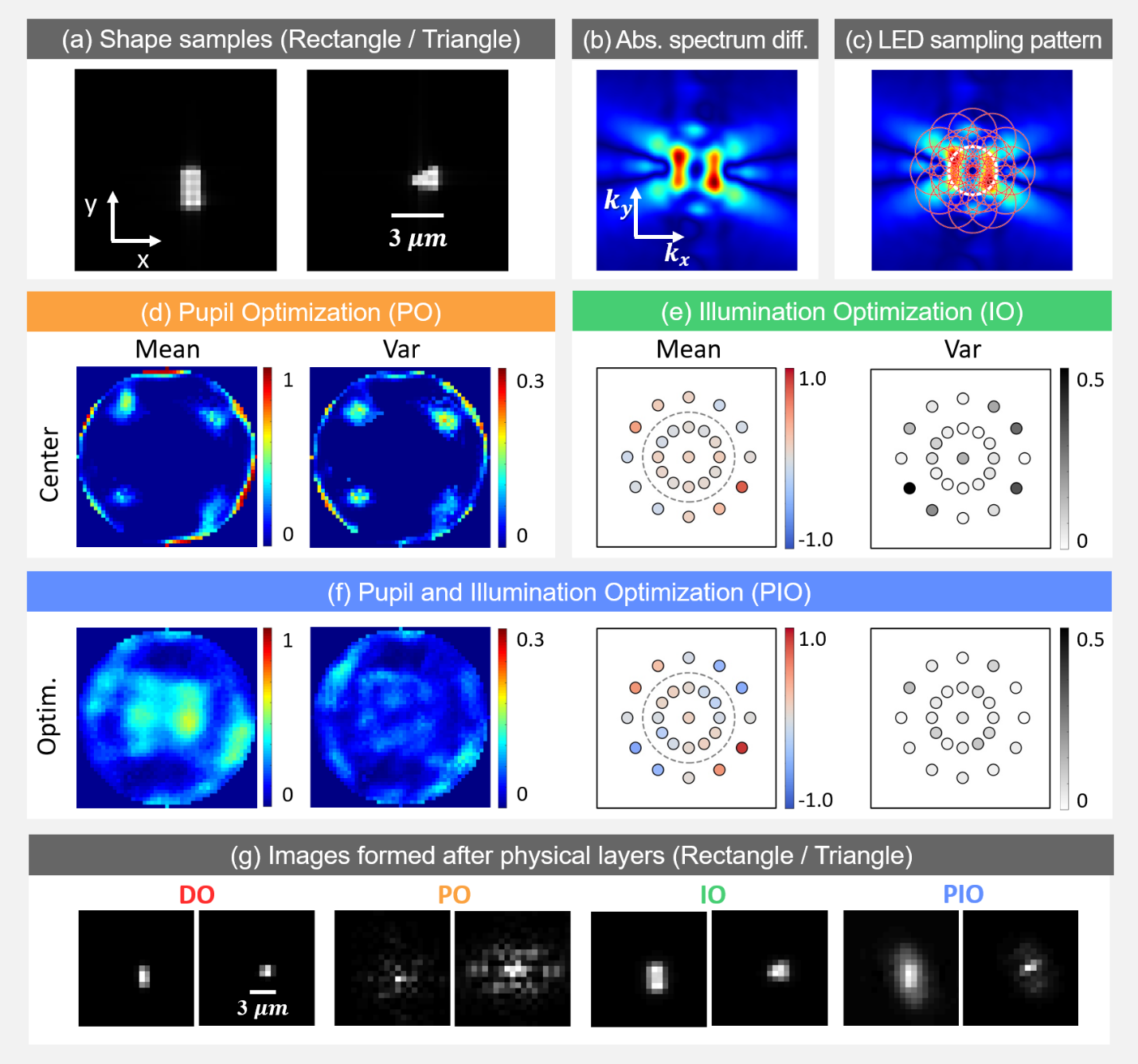
Fig. 2: Simulation results. (a) Example rectangular and triangular samples (absorption map). b) Absolute value of average Fourier spectra difference between two sample types. (c) Marked circles represent spatial frequencies sampled by each corresponding LED illumination angle. (d) Mean and variance of pupil optimization results. Trained pupil is thresholded for better contrast before plotting. (e) Illumination optimization results. (f) Pupil and Illumination optimization results. (g) Example images.
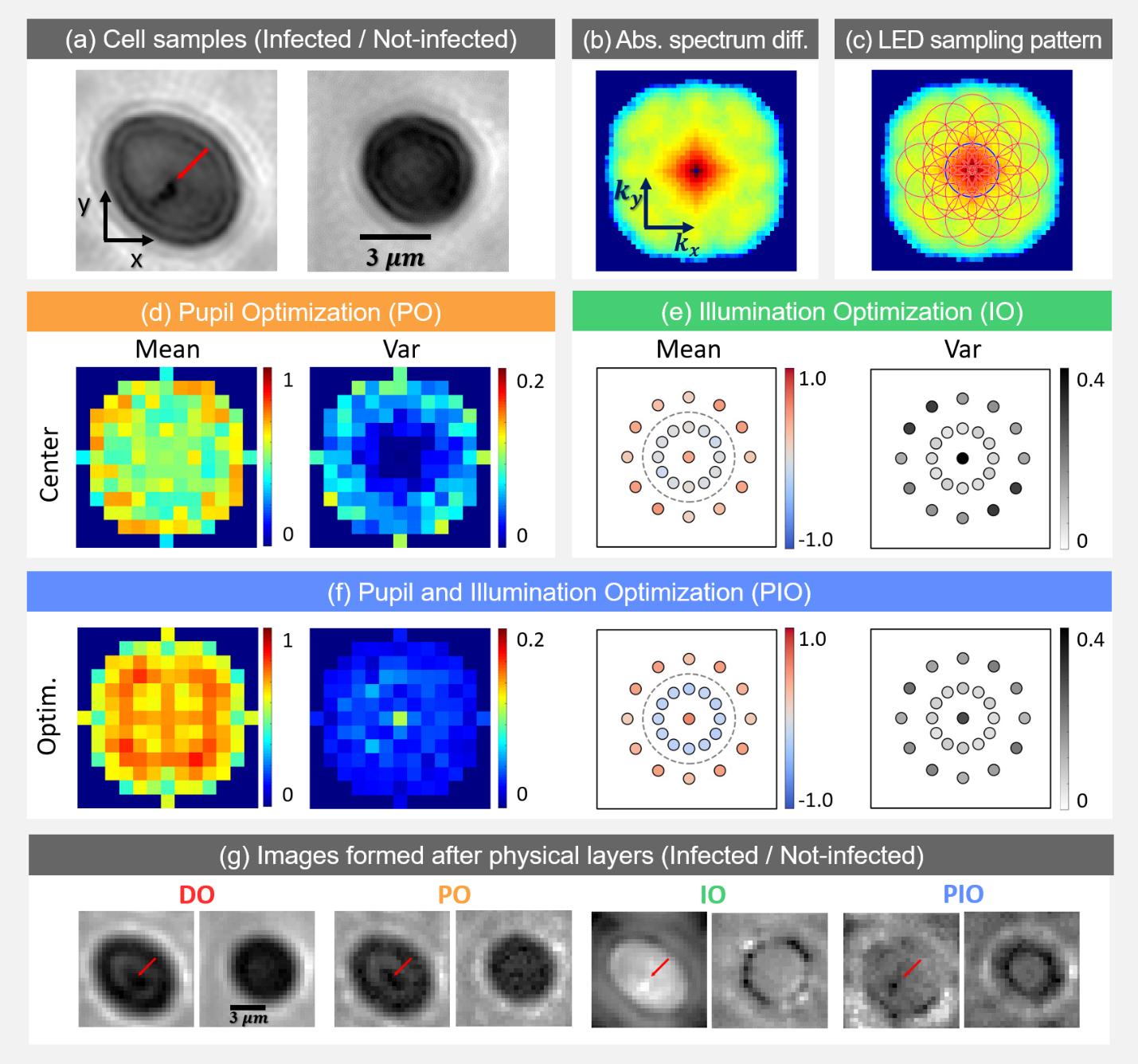
Fig. 3: Malaria parasite detection results. (a) Sample highresolution images of infected (see below) and not-infected cells. (b) Log plot of absolute value of Fourier spectra difference between two cell categories. (c) Each red circle represents spatial frequencies sampled by the corresponding illumination angle. (d) Mean and variance of pupil optimization results. (e) Illumination optimization results. (f) Pupil and Illumination optimization results. (g) Example images.