Alex Muthumbi1,+,Amey Chaware2,+, Kanghyun Kim2, Kevin Zhou3, Pavan Chandra Konda3, Richard Chen4, Benjamin Judkewitz5, Andreas Erdmann6,1, Barbara Kappes7, Roarke Horstmeyer2,3
1: School of Advanced Optical Technologies, Friedrich-Alexander University, Erlangen, Germany.
2: Department of Electrical and Computer Engineering, Duke University, Durham NC, USA.
3: Department of Biomedical Engineering, Duke University, Durham NC, USA.
4: Y Combinator Research, San Francisco, CA.
5: NeuroCure Cluster of Excellence, Charite Universitatsmedizin and Humboldt University, Berlin, Germany.
6: Fraunhofer IISB, Erlangen, Germany.
7: Department of Chemical and Biological Engineering, Friedrich-Alexander University, Erlangen, Germany.
+ denotes equally contributing authors
Links to
paper,
code, and
data
Abstract
Since its invention, the microscope has been optimized for interpretation by a human observer. With the recent development of deep learning algorithms for automated image analysis, there is now a clear need to re-design the microscope's hardware for specific interpretation tasks. To increase the speed and accuracy of automated image classification, this work presents a method to co-optimize how a sample is illuminated in a microscope, along with a pipeline to automatically classify the resulting image, using a deep neural network. By adding a ''physical layer'' to a deep classification network, we are able to jointly optimize for specific illumination patterns that highlight the most important sample features for the particular learning task at hand, which may not be obvious under standard illumination. We demonstrate how our learned sensing approach for illumination design can automatically identify malaria-infected cells with up to 5-10% greater accuracy than standard and alternative microscope lighting designs. We show that this joint hardware-software design procedure generalizes to offer accurate diagnoses for two different blood smear types, and experimentally show how our new procedure can translate across different experimental setups while maintaining high accuracy.
Our Setup
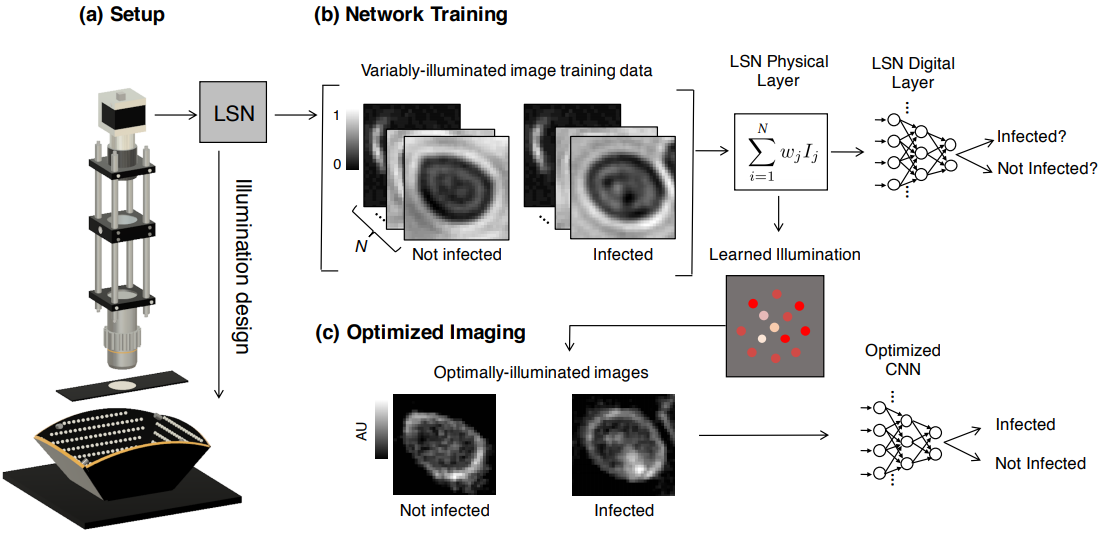
Figure 1: We present a learned sensing network (LSN), which optimizes a microscope's illumination to improve the accuracy of automated image classification. (a) Standard optical microscope outfitted with an array of individually controllable LEDs for illumination. (b) Network training is accomplished with a large number of training image stacks, each containing Ns uniquely illuminated images. The proposed network's physical layer combines images within a stack via a weighted sum before classifying the result, where each weight corresponds to the relative brightness of each LED in the array. (c) After training, the physical layer returns an optimized LED illumination pattern that is displayed on the LED array to improve classification accuracies in subsequent experiments.
Results
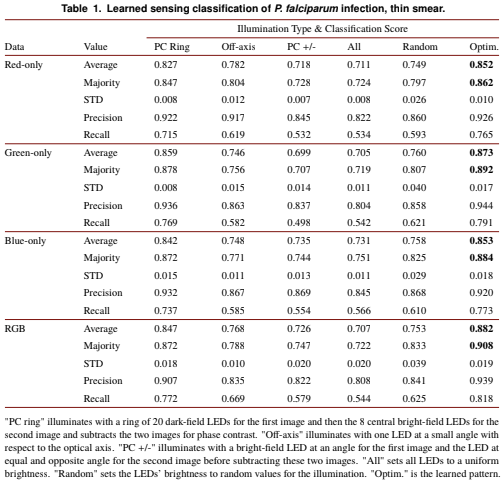
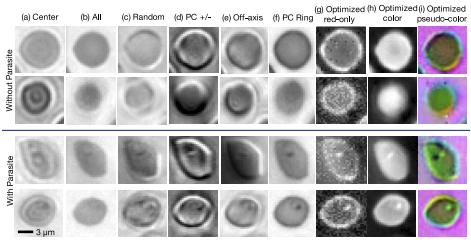
Figure 2: Example images of individual thin-smear blood cells under different forms of illumination. Top two rows are negative examples of cells that do not include a malaria parasite, bottom two rows are positive examples that contain the parasite. Example illuminations include from (a) just the center LED, (b) uniform light from all LEDs, (c) all LEDs with uniformly random brightnesses, (d) a phase contrast-type (PC) arrangement, (e) an off-axis LED, (f) a phase contrast (PC) ring, (g) optimized pattern with red illumination, (h) optimized multispectral pattern, and (i) the same as in (h) but showing response to each individual LED color in pseudo-color, to highlight color illumination’s effect at different locations across the sample.
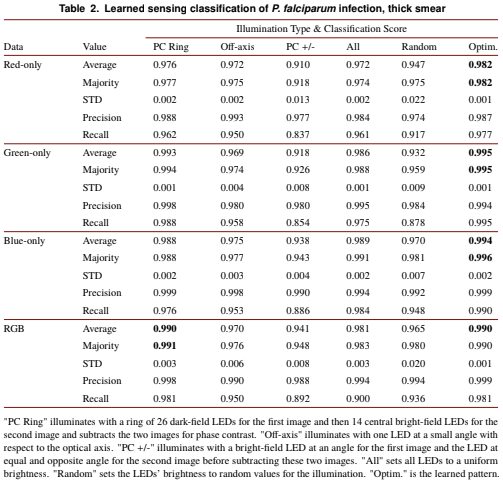
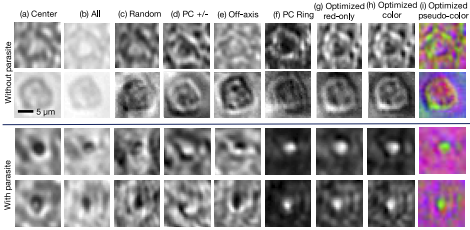
Figure 3: Example images of individual thick-smear blood cells under different forms of illumination. Top two rows are negative examples of cells that do not include a malaria parasite, bottom two rows are positive examples that contain the parasite. Example illuminations include from (a) just the center LED, (b) uniform light from all LEDs, (c) all LEDs with uniformly random brightnesses, (d) a phase contrast-type (PC) arrangement, (e) an off-axis LED, (f) a phase contrast (PC) ring, (g) optimized pattern with red illumination, (h) optimized multispectral pattern, and (i) the same as in (h) but showing response to each individual LED color in pseudo-color, to highlight color illumination’s effect at different locations across the sample.
Code and Data
Code
Data
README




